Article
2 September 2025
Two research voyages off south-eastern Australia provided the opportunity to test and refine environmental DNA (eDNA) sampling for monitoring offshore marine life.
The voyages, conducted in July 2023 and May 2024, were part of the South-East Australian Marine Ecosystem Survey (SEA-MES) on the CSIRO Research Vessel (RV) Investigator. More than 500 eDNA samples were collected and analysed from 91 survey sites along the continental shelf between Tasmania and southern New South Wales. This is the first comprehensive eDNA record for research and comparison in this region of Australia.
A Marine and Coastal Hub project identified thousands of organisms from the eDNA fragments, and compared the results with those of traditional survey methods including fish trawls, plankton sampling, and deep-towed camera deployments. The project built a comprehensive ecological dataset, advanced sampling and analytical approaches, and provided insight into the application of eDNA sampling in marine conservation and management, including for Australian Marine Parks.
“There has been a rapid rise in the use of eDNA approaches to assess and monitor biodiversity for natural resource management,” project leader Dr Bruce Deagle of CSIRO says. “This technology offers the potential to revolutionise environmental monitoring, but widespread adoption still depends on user acceptance and a solid understanding of the insights eDNA data can provide.

“Our initial exploration of several eDNA datasets has demonstrated the exciting potential of eDNA-derived biodiversity information. We hope these will form the basis of detailed case studies and provide reference points for future longitudinal research.”
Dr Bruce Deagle, CSIRO
Evaluating eDNA survey methods
The research used DNA metabarcoding: a powerful technique that analyses tiny fragments of DNA left behind in the environment by marine life. The fragments are amplified, sequenced and matched to known marine taxa using existing reference databases. Like a biological fingerprint, it allows scientist to identify and catalogue species from a simple water sample.
Two distinct approaches were employed to collect eDNA on the SEA-MES surveys:
- Active sampling: considered the conventional approach, this method utilised vacuum pumps to filter water samples collected at specific depths in the water column via a CTD rosette, capturing eDNA onto fine membranes.
- Passive sampling: a novel, CSIRO-developed sampler was mounted on a deep-towed camera system. This device collected an integrated water sample across a horizontal transect, providing a continuous record of near-bottom biodiversity.
The fish diversity measured with each of these eDNA collection methods was similar. It is expected that the passive sampling can be further optimised to increase the eDNA information obtained from the much large volume of water sampled with this method.
Overall patterns of biodiversity in the eDNA samples revealed differences in community composition between surface and bottom collected eDNA. For example, on average, twice as many fish species were detected near the seafloor than in the surface water. Patterns also emerged separating fish communities by depth of the bottom and by latitude.
Comparison with bottom trawling
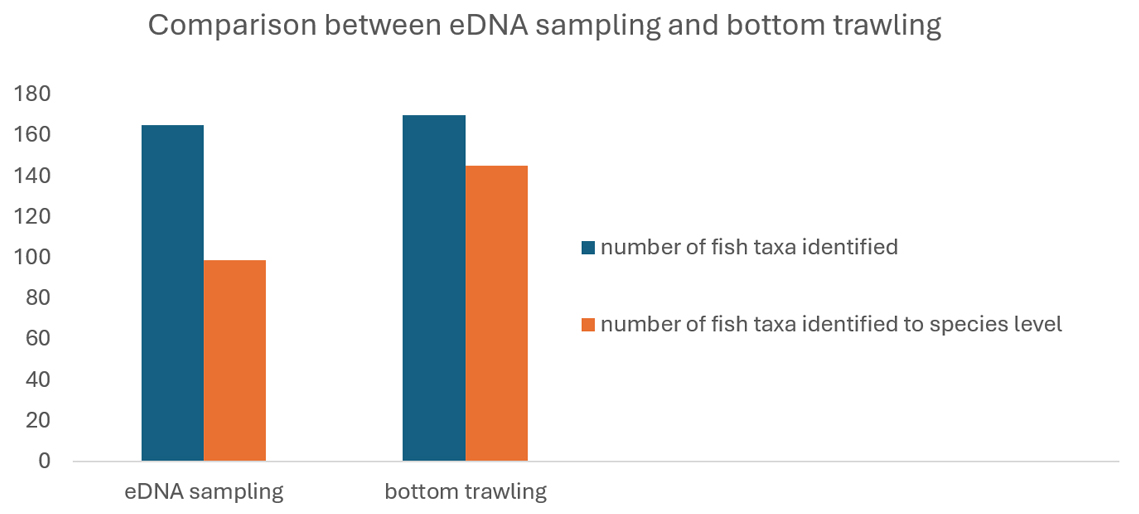
At survey sites where trawling and eDNA sampling methods were paired, the trawling identified slightly more taxa and had better taxonomic resolution which allowed more species-level identifications. Trawling identified 170 fish taxa (145 at the species level), while eDNA sampling identified 165 taxa (99 at the species level).
Of the top 20 taxa identified by each sampling method (based on mass of fish in trawl and proportion of eDNA sequences recovered), nine fish were found in both lists. A notable difference, however, was the dominance of shark and ray species biomass in the trawl catch, while none of these species appeared in eDNA top 20. Conversely, the eDNA data had a large proportion of sequence reads from several small schooling fish that were not dominant in trawl catch.
This complementarity suggests that a combination of methods provides the most comprehensive biodiversity assessment. The main benefits of eDNA biodiversity data are that that it can be produced without the negative impacts of bottom trawling, and it can be carried out in habitats where trawling is not possible (such as rocky reefs, steep slopes). The eDNA data produced here in conjunction with trawl data will allow the utility of eDNA data to be evaluated by marine managers.



Patterns in plankton
Plankton eDNA was another focus, and it revealed a clear connection between the microscopic world and the larger marine community. In May 2024, when nutrient-rich waters were upwelled onto the shelf, copepods dominated the plankton in shallower areas, while deeper slope waters were rich in Radiolaria but low in phytoplankton. These patterns were consistent with the spatial distribution of fish eDNA, suggesting links between plankton structure and fish diversity. A Radiolaria-dominated plankton assembly appeared to negatively impact the diversity of fish present on the deeper (>100 metre) continental slope.
Tree of Life eDNA metabarcoding
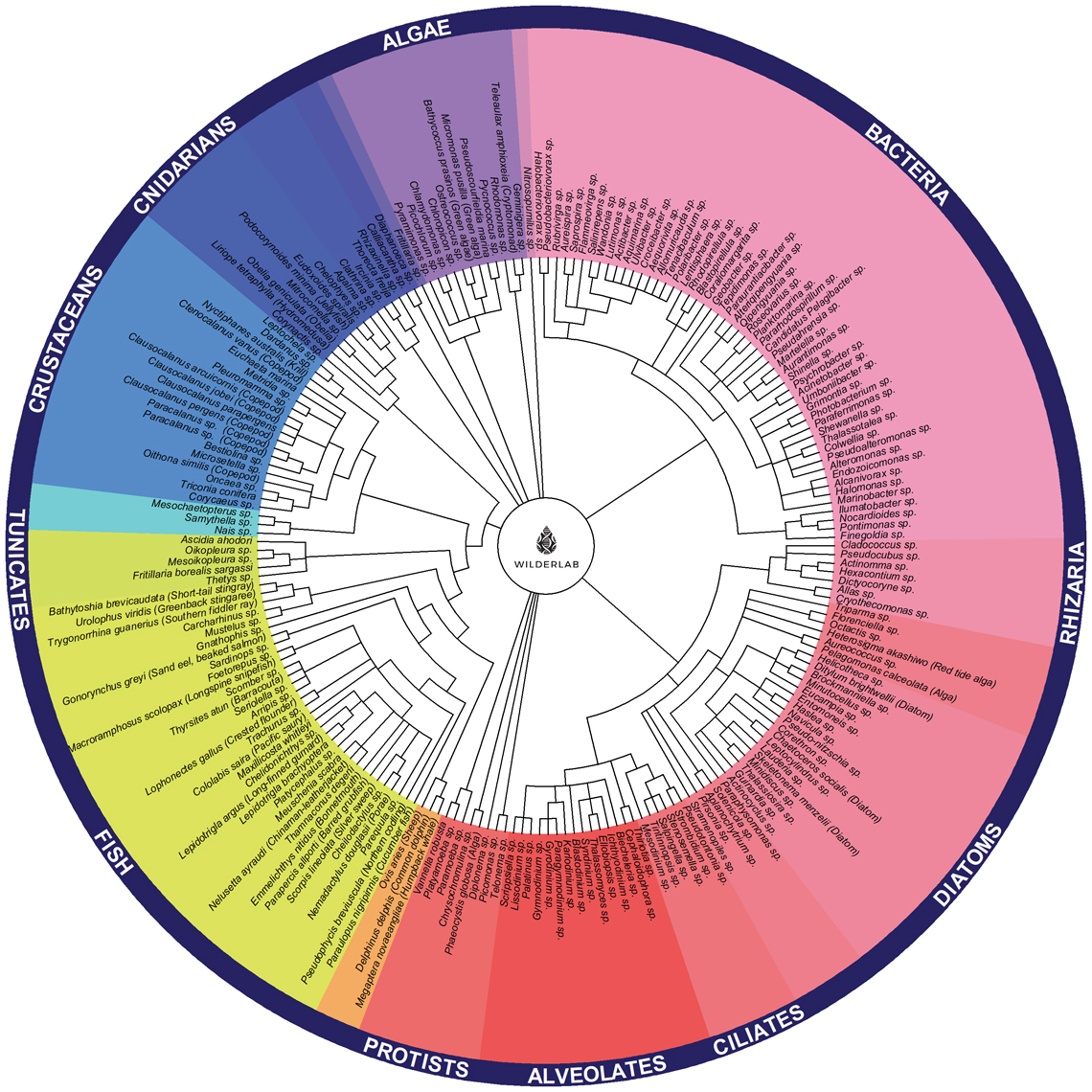
The project also investigated the use of Tree of Life (ToL) eDNA metabarcoding, an approach that combines multiple genetic assays to provide a broad, high-resolution overview of the phylogenetic diversity within a sample, from fish and crustaceans to bacteria. The promising results indicate that ToL metabarcoding is an effective tool for maximising information from a single eDNA sample, offering a powerful capability for rapid biological characterisation.
“There has been a rapid rise in the use of eDNA approaches to assess and monitor biodiversity for natural resource management,” Dr Deagle says. “This technology offers the potential to revolutionise environmental monitoring, but widespread adoption still depends on user acceptance and a solid understanding of the insights eDNA data can provide. This project provides several detailed eDNA case studies and reference points for future longitudinal research.”
Further information
- Technical report: Environmental DNA for measuring offshore marine biodiversity: what can DNA in water collected from the RV Investigator tell us?
Acknowledgement
This research was supported by grants of sea time on RV Investigator from the CSIRO Marine National Facility, which is supported by funding from the Australian Government’s National Collaborative Research Infrastructure Strategy.
The Southeast Australian Marine Ecosystem Survey (SEA-MES) is a multi-year, interdisciplinary project that is revisiting previous bio-physical and ecosystem surveys of the Australian southeast continental shelf, and documenting changes in the 25 years since it was last examined. It is also establishing a baseline for the continental slope. The SEA-MES mission is to understand what has changed, what contribution has human activity made, and what is the prospect for the future.